Akshay R. Yadav1*, Shital B. Khade2, Vaibhavi Annasaheb Soundatti3
1Assistant Professor, Department of Pharmaceutical Chemistry, Rajarambapu College of Pharmacy, Kasegaon, Dist- Sangli, Maharashtra, India-415404
2Department of Pharmaceutics, Rajarambapu College of Pharmacy, Kasegaon, Dist- Sangli, Maharashtra, India-415404
3Department of Pharmaceutics, Bharati Vidyapeeth College of Pharmacy, Kolhapur, Maharashtra, India-416013
ABSTRACT
Morpholine is a heterocycle used in a variety of drugs, both approved and experimental, as well as bioactive molecules. The morpholine ring is a versatile and readily available synthetic building block that can be used as an amine reagent or built using a variety of synthetic methodologies. Molecular docking studies against Structure of PanDDA analysis group deposition-Crystal Structure of COVID-19 main protease in complex with Z219104216 (PDB code- 5R82) and also the activities are compared with FDA-approved few human trial drugs such as hydroxychloroquine. To identify the hypothetical binding motif of the title compounds using VLifeMDS software by in-silico (molecular docking studies). These findings showed that the binding energy in all active compounds ranged from -20.55 to -74.55 kcal/mol. If compared to the standard -80.12 kcal/mol). Compound code 2b and 2e were found to be potent with a docking score of -74.55 and -60.29 respectively. As the world’s population increases and health problems expand accordingly, need to discover new therapeutics will become even more diring. There are several examples of morpholine bioactive molecular targets in which the morpholine moiety has been shown to play a significant role; it is an important component of the pharmacophore for certain enzyme active site inhibitors, and it confers selective affinity for a wide range of receptors.
Keywords: Morpholine, Molecular docking, SARS-CoV-2 inhibitors, Hydroxychloroquine.
INTRODUCTION
Several interesting biologically active compounds containing substituted morpholine units have been discovered recently. Despite the medicinal value of these molecules, new approaches to their synthesis have yet to be developed1. As on 21st September 2020; COVID-19 (novel RNA virus) has infected >31 million individuals and caused approximately 1 million global deaths. The novel human RNA virus is subjected to the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) which primarily gains entrance to cells via binding of SARS-CoV-2 Spike glycoprotein to angiotensin converting enzyme 2 (ACE-2) and subsequent endocytosis2-5. Scholars have made careful attention to nitrogen-containing heterocycles in recent decades due to their high therapeutic potential. Both natural or synthetic, they are commonly used as key components in biological processes due to their interesting biological properties. Quinine, ellipticine, theophylline, emetine, papaverine, procaine, codeine, and morphine are only a few of the nitrogen-containing heterocycles that have made an indelible mark as phytochemical drugs in the plant kingdom6. Apart from the wide distribution of nitrogen-containing heterocycles in natural products, they also play an important role in biochemical processes in living cells, with aromatic heterocycles constituting the majority of enzymes and non-amino acids constituting the majority of coenzymes. Some essential vitamins are built on aromatic heterocyclic scaffolds, and moieties are aromatic nitrogen heterocycles. New methods for the synthesis of nitrogen-containing heterocycles, which are usually pharmacophoric fragments or naturally biologically active organic compounds, have recently received a lot of publicity. In general, new trends in this field of chemistry will arise from the development of new schemes for the creation of heterocycles as well as the synthesis of unique and readily available starting compounds capable of specific transformation pathways into the desired nitrogen-containing heterocycles7. Growth of green chemistry holds necessary potential for the reduction of by product, a reduction in the waste production and a lowering of energy costs. Due to its ability to couple directly with reaction molecule and passing thermal conductivity leading to fast rise in the temperature microwave irradiation had used to improve many organic synthesis8-18. The Principle behind the heating in microwave oven is because interaction of charged particle of reaction material with electromagnetic wavelength of particular frequency19-28. The phenomena of the producing heat by electromagnetic irradiation are either by conduction or collision. The application of green chemistry principles and practices renders regulation, control, clean-up, and remediation of the environment29-38. Because of ADME failure, it is important to conduct docking studies before pharmacological activity, as it is simple to predict the probable pharmacological activity by receptors with the help of structure of compounds. In the discovery of effective medicines for prevention and treatment, an outbreak of coronavirus disease (COVID-19) caused by the novel extreme acute respiratory syndrome coronavirus-2 (SARS-CoV-2) poses an unprecedented obstacle. Given the rapid pace of scientific research and clinical data provided by the large number of people who are rapidly infected with SARS-CoV-2, clinicians need reliable evidence of good medical care for this infection, as it is simple to do in-silico analysis in the initial stage with the aid of molecular docking software with help of chemical structure of compound39-56. It is necessary to enhance both enzymatic stability and membrane permeation in the formulating drug delivery system for protein and peptide drugs. Soon, someday, you might be making your own drugs at home. That is because researchers have adapted a 3D printer from basic, readily available medicinal active agents fed into a drug delivery system57-59. Molecular docking is an appealing scaffold for understanding medicinal biomolecular interactions in rational drug design as well as in the mechanical analysis in order, primarily noncovalently, to insert the molecule (ligand) into the favorite binders of the particular target area of the DNA/protein (receptor). The information gathered from the docking method can be used to demonstrate the binding energy, free energy and complex stability. The docking are currently used to forecast the preliminary ligand-receptor complex binding parameters. The main user interface continues to be expanded by commercial software programs. In the high end packages, new algorithms from industry and academia are easily implemented. Public domain packages are becoming more stable and deliver functionality that continues to double in speed every year and half computers surpassing some of the commercial offerings, while graphic displays have become more sophisticated and intuitive. All these components make molecular docking an important part of the design of drugs. In exciting new techniques such as computational enzymology, genomics, and proteomic search engines, its function continues to be expanded60-61.
MATERIALS AND METHODS
Molecular Docking Study
The VLifeMDS 4.1 software was used to perform the molecular docking study. There are all six 1,2-diphenyl-1H-benzimidazole products. VLifeMDS 4.1 software has provided both rigid (no torsional flexibility for both a protein and a ligand) and flexible (torsional flexibility for a rigid protein ligand) molecular docking. Either experimentally known or theoretically developed through knowledge-based protein modeling or homology modeling was the target or receptor. The molecular docking tool was designed to obtain a preferred interaction geometry of ligand-receptor complexes with minimum interaction energy assisted by various scoring functions. Electrostatics only, the steric and electrostatic sum (force-field parameters), and the dock score. For lead optimization, this utility allowed us to screen a collection of compounds. The interaction energy between the ligand and the receptor protein is minimized using VLifeMDS62.
Protein Preparation
PanDDA analysis group deposition-Crystal Structure of COVID-19 main protease in complex with Z219104216 (PDB code- 5R82)
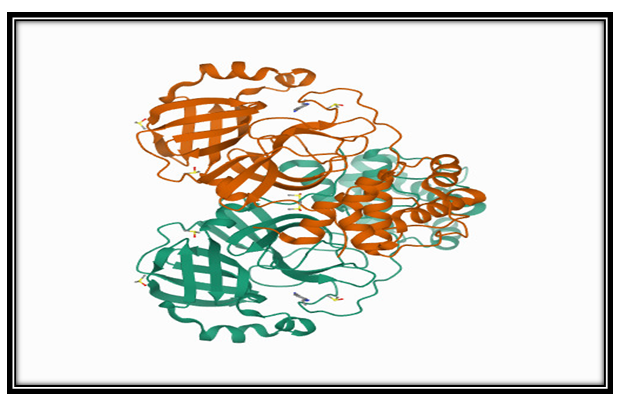
Fig-1: 3D View of Structure of PanDDA analysis group deposition-Crystal Structure of COVID-19 main protease in complex with Z219104216 (PDB code- 5R82)
Ligand preparation
The 2D structures of the compounds were built and then converted into 3D. Then, using MMFF, the 3D structures were energetically minimized to the rms gradient of 0.0163.
Identification of cavities
Enzyme cavities were calculated by using the software’s cavity determination option. To assign an appropriate active site, the cavities in the receptor were mapped. The basic function used to map the cavities was the receptor’s surface mapping and the geometric voids were defined and the void was scaled for its hydrophobic characteristics. Therefore, based on their size and hydrophobic surface area, all the cavities that are present in the receptor are classified and ranked. Considering the dimensions and the hydrophobic surface area, as an active site, the cavity is considered to be the best void64.
Scoring function
The distinction is based on the scoring or fitness feature of good or bad docked conformation. Only electrostatic and both steric and electrostatic interactions between receptor ligand and dock score scoring function are used by MDS fitness functions. The dock score measures binding affinity with a recognized 3D structure of a given protein-ligand complex65-66.
Table 1: Test compounds used in study
| Sr. No | Compound code | Name of compound |
| 1 | 2a | 2-Amino-4-(4-(2-morpholinoethoxy)phenyl)-5-oxo-5,6,7,8-tetrahydro-4H-chromene-3-carbonitrile |
| 2 | 2b | 2-Amino-7,7-dimethyl-4-(4-(2-morpholinoethoxy)phenyl)-5-oxo-5,6,7,8-tetrahydro-4H-chromene-3-carbonitrile |
| 3 | 2c | 2-Amino-7-methyl-4-(4-(2-morpholinoethoxy)phenyl)-5-oxo-4H,5H-pyrano[4,3-b]pyran-3-carbonitrile |
| 4 | 2d | 2-Amino-4-(4-(2-morpholinoethoxy)phenyl)-5-oxo-4H,5H-pyrano[3,2-c]chromene-3-carbonitrile |
| 5 | 2e | 2-Amino-5-oxo-4-(4-(2-(piperidin-1-yl)ethoxy)phenyl)-4H,5H-pyrano[3,2-c]chromene-3-carbonitrile |
| 6 | 2f | 2-Amino-5-oxo-4-(4-(2-(piperidin-1-yl)ethoxy)phenyl)-5,6,7,8-tetrahydro-4H-chromene-3-carbonitrile |
RESULTS AND DISCUSSION
Molecular docking study were subjected on receptor of PanDDA analysis group deposition-Crystal Structure of COVID-19 main protease in complex with Z219104216 (PDB code- 5R82). The compound code (2a-f) shown in the table and the compound code 2b and 2e minimum dock score were found to be potent, with a docking score of -74.55 and -60.29 respectively. Where the main interaction between ligand and receptor can be observed, the best pose obtained by docking results is reported. At the binding pocket, all designed compounds follow a very similar conformation, showing interaction of hydrogen bond with amino acids of GLN1719 aromatic interaction with amino acids of MET165, ASP159, MET49, HIE164, ASN142, ASN15B, ASN119 and ASN142. The standard dock score was found to be -80.12.
Table 2: Docking score of Morpholine derivatives by using GRIP Batch docking.
| Compound code | Name of compound | Docking score (Kcal/mol) |
| 2a | 2-Amino-4-(4-(2-morpholinoethoxy)phenyl)-5-oxo-5,6,7,8-tetrahydro-4H-chromene-3-carbonitrile | -44.19 |
| 2b | 2-Amino-7,7-dimethyl-4-(4-(2-morpholinoethoxy)phenyl)-5-oxo-5,6,7,8-tetrahydro-4H-chromene-3-carbonitrile | -74.55 |
| 2c | 2-Amino-7-methyl-4-(4-(2-morpholinoethoxy)phenyl)-5-oxo-4H,5H-pyrano[4,3-b]pyran-3-carbonitrile | -30.42 |
| 2d | 2-Amino-4-(4-(2-morpholinoethoxy)phenyl)-5-oxo-4H,5H-pyrano[3,2-c]chromene-3-carbonitrile | -20.55 |
| 2e | 2-Amino-5-oxo-4-(4-(2-(piperidin-1-yl)ethoxy)phenyl)-4H,5H-pyrano[3,2-c]chromene-3-carbonitrile | -60.29 |
| 2f | 2-Amino-5-oxo-4-(4-(2-(piperidin-1-yl)ethoxy)phenyl)-5,6,7,8-tetrahydro-4H-chromene-3-carbonitrile | -41.28 |
| Standard | Hydroxychloroquine | -80.12 |
CONCLUSION
In conclusion, molecular docking studies further assist in understanding in detail the different interactions between the ligands and enzyme active sites and thus assist in developing new potent inhibitors. For all the synthesized compounds, the docking experiments were carried out and the docking score was compared with the Hydroxychloroquine reference compound. The compounds code 1d showed higher binding score.
REFERENCES
- Panneerselvan P, Rajasree R, Vijayalakshmi G, Subramanian E, Sridhar S. Synthesis of Schiff bases of 4-(4-aminophenyl)-morpholineas potential antimicrobial agents. Eur J Med Chem. 2005; 40: 225-229.
- Yadav A, Mohite S. A Review on severe acute respiratory infection (SARI) and its clinical management in suspect/confirmed novel coronavirus (nCoV) cases J. Pharma. Dosage Forms and Tech. 2020; 12(3): 178-180.
- Yadav A, Mohite S. A Review on Novel Coronavirus (COVID-19). International Journal of Pharma Sciences and Research. 2020; 11(5): 74-76.
- Yadav A, Mohite S. Role of Indian Youth in Keeping COVID-19 at Bay. Int J Sci Res Chemi. 2020; 5(5): 46-50.
- Yadav A, Mohite S. A Novel approach for treatment of COVID-19 with Convalescent Plasma. J. Pharma. Dosage Forms and Tech. 2020; 12(3): 227-230.
- Manssour Fraga, C.A.; Barreiro, E.J. New insights for multifactorial disease therapy: The challenge of the symbiotic drugs. Curr. Drug Ther. 2008; 3: 1–13.
- Kimura, I. Medical benefits of using natural compounds and their derivatives having multiple pharmacological actions. Yakugaku Zasshi 2006; 126: 133–143.
- Giguere R, Bray T, Duncan S, Majetich G. Application of commercial microwave ovens to organic synthesis. Tetrahedron Lett. 1986; 27: 4945-4948.
- Rode P, Yadav A, Chitruk A, Mohite S, Magdum C. Microwave assisted synthesis, toxicological assessment using brine shrimp lethality assay and antimicrobial potential of new series of benzimidazole derivatives. Int. J. Curr. Adv. Res. 2020; 09(08)(A): 22900-22905.
- Rajput M. D, Yadav A. R, Mohite S. K. Synthesis, Characterization of Benzimidazole Derivatives as Potent Antimicrobial Agents. International Journal of Pharmacy & Pharmaceutical Research. 2020; 17(4): 279-285.
- Yadav A, Mohite S. Cancer- A Silent Killer: An Overview. Asian J. Pharm. Res. 2020; 10(3): 213-216.
- Yadav A, Mohite S. Antioxidant Activity of Malvastrum Coromandelianum Leaf extracts. Research J. Topical and Cosmetic Sci. 2020; 11(2): 59-61.
- Yadav A, Patil S, Dharanguttikar V, Mohite S. Anthelmintic Activity of Malvastrum Coromandelianum Leaf Extracts against Pheretima Posthuma and Ascardia Galli. Int J Sci Res Chemi. 2020; 5(6): 18-24.
- Yadav A, Mohite S. Anthelmintic and Antibacterial Activity of Psidium Guajava Leaf Extracts. Int J Sci Res Chemi. 2020; 5(6): 06-11.
- Yadav A, Dange V, Mohite S. Pathogensis of Cell Injury. Int J Sci Res Chemi. 2020; 5(6): 12-18.
- Suryawanshi V, Yadav A, Birajdar R, Jagtap N, Vambhurkar G, Patil P. Optimization of ayurvedic herbal medicine by nanoformulation. Asian J. Res. Pharm. Sci. 2019; 9(1): 55-56.
- Yadav A, Honmane P, Bhosale M, Chitruk A, Rode P, Birajdar R, Rajput M, Suryawanshi V, Patil S, Patil, Jagtap N, Mohite S, Dange V, Vambhurkar G. Antifungal Activity of Malvastrum Coromandelianum Leaf Extracts. International Journal of Scientific Research in Chemistry. 2020; 5(6): 01-05.
- Yadav A, Mohite S. Screening of In-vitro anti-inflammatory and Antifungal assay of Psidium guajava Leaf Extracts. Research J. Topical and Cosmetic Sci. 2020; 11(2): 62-64.
- Yadav A, Mohite S. Rajput M, Suryawanshi V, Birajdar R, Patil M. Antioxidant Activity of Psidium guajava Leaf Extracts. Res. J. Pharma. Dosage Forms and Tech. 2020; 12(3): 159-161.
- Yadav A, Mohite S. A Review on Zika Virus Infection. J. Pharma. Dosage Forms and Tech. 2020; 12(4): 245-249.
- Yadav A, Mohite S. Toxicоlogical Evaluation of Psidium guajava Leaf Extracts using Brine Shrimp (Artemia salina L.) Model. J. Pharma. Dosage Forms and Tech. 2020; 12(4): 198-120.
- Honmane P, Yadav A, Singh S, Mohite S. Synthesis, Characterization and Antiplatelet Activity of Antithrombotic novel 2,5-substituted aryl-7-phenyl-1,3,4-oxadiazolo-[3,2-a]-1,3,5-triazine Derivatives. Journal of University of Shanghai for Science and Technology. 2020; 22(11): 881-898.
- Patil S, Yadav A, Chopade A, Mohite S. Design, Development and Evaluation of Herbal Mouthwash for Antibacterial Potency against Oral Bacteria. Journal of University of Shanghai for Science and Technology. 2020; 22(11): 881-898.1137-1148.
- Honmane P, Yadav A, Singh S, Mohite S. Synthesis of Pyrazole Acrylic acid based Oxadiazole and Amide Derivatives as Larvicidal and Antitubercular agents. Seybold Rep. 2020; 25(10): 516-530.
- Yadav A, Mohite S. Recent Advances in the Ultrasound-Assisted Synthesis of Oxadiazole and Thiazole Derivatives. Res. J. Pharma. Dosage Forms and Tech. 2020; 12(4): 225-228.
- Yadav A, Mohite S. An Overview on Ebola Virus Disease. Res. J. Pharma. Dosage Forms and Tech.2020; 12(4): 230-235.
- Yadav A, Mohite S. Carbon Nanotubes as an Effective Solution for Cancer Therapy. Res. J. Pharma. Dosage Forms and Tech. 2020; 12(4): 238-241.
- Honmane P, Yadav A, Singh S, Mohite S. 3D printing technology in pharmaceuticals and biomedical. World J Pharm Pharm Sci. 2020; 9(9): 598-609
- Gavali K, Yadav A, Howal R, Tamboli A. Preliminary Phytochemical Screening and HPTLC Finger printing of Leaf Extracts of Tectona grandis Linn Journal of University of Shanghai for Science and Technology. 2020; 22(11): 1804-1815.
- Dange V, Dinde S, Doiphode A, Dhavane S, Dudhal B, Shid S, Yadav A. Formulation and Evaluation of Herbal gel Containing Lantana Camara for Management of Acne Vulgaris. Journal of University of Shanghai for Science and Technology.2020; 22(11): 799-809.
- Yadav A, Mohite S. Screening of In-vitro anti-inflammatory and Antibacterial assay of Malvastrum Coromandelianum. International Journal of Pharma Sciences and Research. 2020; 11(4): 68-70.
- Suryawanshi V, Yadav A, Mohite S. Toxicological Assessment using Brine Shrimp Lethality Assay and Antimicrobial activity of Capparis Grandis. Journal of University of Shanghai for Science and Technology. 2020; 22(11): 746-759.
- Pathade K, Mohite S, Yadav A. 3D-QSAR And ADMET Prediction Of Triazine Derivatives For Designing Potent Anticancer Agents. Journal of University of Shanghai for Science and Technology. 2020; 22(11): 1816-1833.
- Yadav A, Dange V. Biochemical Mediators of Inflammation and Basic Principles of Wound Healing in the Skin. International Journal of Pharmacology and Pharmaceutical Sciences. 2020; 2(1): 14-18.
- Yadav A, Dange V. Mechanism Involved in the Process of Inflammation. International Journal of Pharmacology and Pharmaceutical Sciences. 2020; 2(1): 01-06.
- Yadav A, Mohite S. Phytochemical and Pharmacological Review of Embelia ribes. Int J Sci Res Chemi. 5(5): 57-62.
- Pawara N, Yadav A, Mohite S. Pharmacognostic, Phytochemical Investigation and Antioxidant Potential of Embelia ribes. Int J Sci Res Chemi. 5(6): 27-34.
- Yadav A, Mohite S. Pharmaceutical Process Scale-Up. Int J Sci Res Chemi. 5(5): 49-55
- Rajput M, Yadav A. Green Chemistry Approach for Synthesis of Some 1,3,4-Oxadiazole Derivatives As Potent Antimalarial Agents Journal of University of Shanghai for Science and Technology. 2020; 22(11): 1854-1869.
- Yadav A, Mohite S. Transforming Global Health. Int J Sci Res Chemi. 5(6): 41-48.
- Yadav A, Rajput M, Gavali K, Mohite S. In-vitro Hypoglycemic Activity of Barleria prionitis L. Int J Sci Res Chemi. 5(5): 63-70.
- Yadav A. Transition-metal and Organo-catalysis in Organic Synthesis: Metal-catalyzed Reactions. International Journal of Pharmacy and Pharmaceutical 2020; 2(1): 06-09.
- Yadav A. Role of Medicinal Chemistry in the Current Scenario. International Journal of Pharmacy and Pharmaceutical 2020; 2(1): 27-36
- Yadav A, Honamane P, Rajput M, Dange V, Salunkhe K, Kane S, Mohite S. Antimalarial Activity of Psidium guajava Leaf Extracts. Int J Sci Res Chemi. 2020; 5(6): 63-68.
- Honmane P, Yadav A, Singh S, Mohite S. Microwave Assisted Synthesis of Novel Benzimidazole Derivatives as Potent Antileishmanial and Antimalarial Agents. Int. J. Curr. Adv. Res. 2020; 09(07)(B): 22742-22746.
- Yadav A, Mohite S. Design, synthesis and characterization of some novel benzamide derivatives and it’s pharmacological screening. Int. j. sci. res. sci. technol. 2020; 7(2), 68-74.
- Chitruk A, Yadav A Rode P, Mohite S, Magdum C. Microwave assisted synthesis, antimicrobial and anti-inflammatory potential of some novel 1,2,4-triazole derivatives. Int. j. sci. res. sci. technol. 2020; 7(4): 360-367.
- Yadav A, Mohite S. Anticancer Activity and In-Silico ADMET Analysis of Malvastrum Coromandelianum. International Journal of Pharma Sciences and Research. 2020; 11(5): 71-73.
- Yadav A, Mohite S. In-Silico ADME Analysis of 1, 3, 4-oxadiazole derivatives as CDK9 Inhibitors. International Journal of Chemical Science. 2020; 4(3): 01-04.
- Yadav A, Mohite S. ADME analysis of phytochemical constituents of Psidium guajava. Asian J. Res. Chem. 2020; 13(5): 373-375.
- Yadav A, Mohite S. Prediction and Optimization of Drug Metabolism and Pharmacokinetics Properties Including Absorption, Distribution, Metabolism, Excretion, and the Potential for Toxicity Properties. Int J Sci Res Chemi. 2020; 4(5): 47-58.
- Yadav A, Mohite S. Molecular Properties Prediction and Synthesis of Novel Benzimidazole Analogues as Potent Antimicrobial agents. Int J Sci Res Chemi. 2019; 4(6): 23-34.
- Chitruk A, Yadav A, Rode P, Mohite S, Magdum C. Microwave assisted synthesis, antimicrobial and anti-inflammatory potential of some novel 1,2,4-triazole derivatives. Int. j. sci. res. sci. technol. 2020; 7(4): 360-367.
- Yadav A, Mohite S. Production of Statins by Fungal Fermentation. Int J Sci Res Chemi. 2020; 5(1): 59-64.
- Yadav A, Gavali K, Rajput M, Pathade K, Patil S, Dharanguttikar V, Mohite S. Anthelmintic Activity of Malvastrum Coromandelianum Leaf Extracts against Pheretima Posthuma and Ascardia Galli. Int J Sci Res Chemi. 2020; 5(6): 18-24.
- Yadav A, Rajput M, Gavali K, Pathade K, Honmane P, Mohite S. Anthelmintic and Antibacterial Activity of Psidium Guajava Leaf Extracts. Int J Sci Res Chemi. 2020; 5(6): 6-11.
- Yadav A, Mohite S. Toxicоlogical Evaluation of Eclipta alba using Brine Shrimp (Artemia salina L.) Model. Int J Sci Res Chemi. 2020; 5(6): 56-62.
- Yadav A, Mohite S. QSAR Studies as Strategic Approach in Drug Discovery. Int J Sci Res Chemi. 2019; 4(6): 16-22.
- Rode P, Yadav A, Chitruk A, Mohite S, Magdum C. Synthesis, Anticancer and Molecular Docking Studies of N-(1H-benzimidazol-2-yl-carbamothioyl)benzamide Analogues. Int. j. sci. res. sci. technol. 2020; 5(6): 204-212.
- Bhosale M, Yadav A, Magdum C, Mohite S. Molecular Docking Studies, Synthesis, Toxicological Evaluation using Brine Shrimp (Artemia salina L.) Model and Anti-inflammatory Activity of Some N-(substituted)-5-phenyl-1,3,4-thiadiazol-2-amine Derivatives. Int J Sci Res Sci & Technol. 2020; 7(5): 51-62.
- Bhosale M, Yadav A, Magdum C, Mohite S. Microwave Assisted Synthesis, Molecular Docking Studies and Anticancer Screening of Some 1,3,4-thiadiazole Derivatives. Journal of University of Shanghai for Science and Technology.2020; 22(11):520-534.
- Birajdar R, Yadav A, Patil S, Chitruk A, Kane S, Mohite S, Magdum C. Pharmacognostic and Phytochemical Investigation, Molecular Docking Studies of Phytoconstituents and Anticancer Potential of Capparis Decidua (Forsk) Edgew. Journal of University of Shanghai for Science and Technology. 2020; 22(11): 500-519.
- Yadav A, Mohite S. Pharmacophore Mapping and Virtual Screening. Int J Sci Res Chemi. 5(5): 77-80.
- Pathade K, Mohite S, Yadav A. Synthesis, Molecular Docking Studies of Novel 4-(Substituted Ph Phenyl Amino)-6-(Substituted Aniline)-N’-Aryl-1,3,5-Triazine-2-Carbahydrazide Derivatives As Potent Antitubercular Agents. Journal of University of Shanghai for Science and Technology. 2020; 22(11): 1891-1909.
- Yadav A, Mohite S. Homology Modeling and Generation of 3D-structure of Protein. J. Pharma. Dosage Forms and Tech. 2020; 12(4): 218-224.
- Bhosale M, Yadav A, Magdum C, Mohite S. Synthesis, molecular docking studies and biological evaluation of 1,3,4-thiadiazole derivatives as antimicrobial agents. I J. Curr. Adv. Res. 2020; 09(08)(A): 22894-22899.
